Coming back to the Boundary element method
During October (2017) I wrote a program per day for some well-known numerical methods in both Python and Julia. It was intended to be an exercise to learn some Julia. You can see a summary here. I succeeded in 30 of the challenges, except for the BEM (Boundary Element Method), where I could not figure out what was wrong that day. The original post is here.
Thomas Klimpel found the mistake and wrote an email where he described my mistakes. So, I am creating a new post with a correct implementation of the BEM.
The Boundary Element Method
We want so solve the equation
with
For this method, we need to use an integral representation of the equation, that, in this case, is
with
and
Then, we can form a system of equations
that we obtain by discretization of the boundary. If we take constant variables over the discretization, the integral can be computed analytically by
and
for points \(n\) and \(m\) in different elements, and the subindices \(A,B\) refer to the endpoints of the evaluation element. We should be careful evaluating this expression since both \(r_A\) and \(r_B\) can be (close to) zero and make it explode. Also, here it was the main problem since I forgot to compute the angles with respect to elements that are, in general, not aligned with horizontal or vertical axes.
For diagonal terms the integrals evaluate to
and
with \(L\) the size of the element.
Following is the code. Keep in mind that this code works for purely Dirichlet problems. For mixed Dirichlet-Neumann the influence matrices would need rearranging to separate known and unknowns in opposite sides of the equation.
You can download the files for this project here. It includes a YML file to create a conda environment with the dependencies listed. For example, it uses version 3.0 of Meshio.
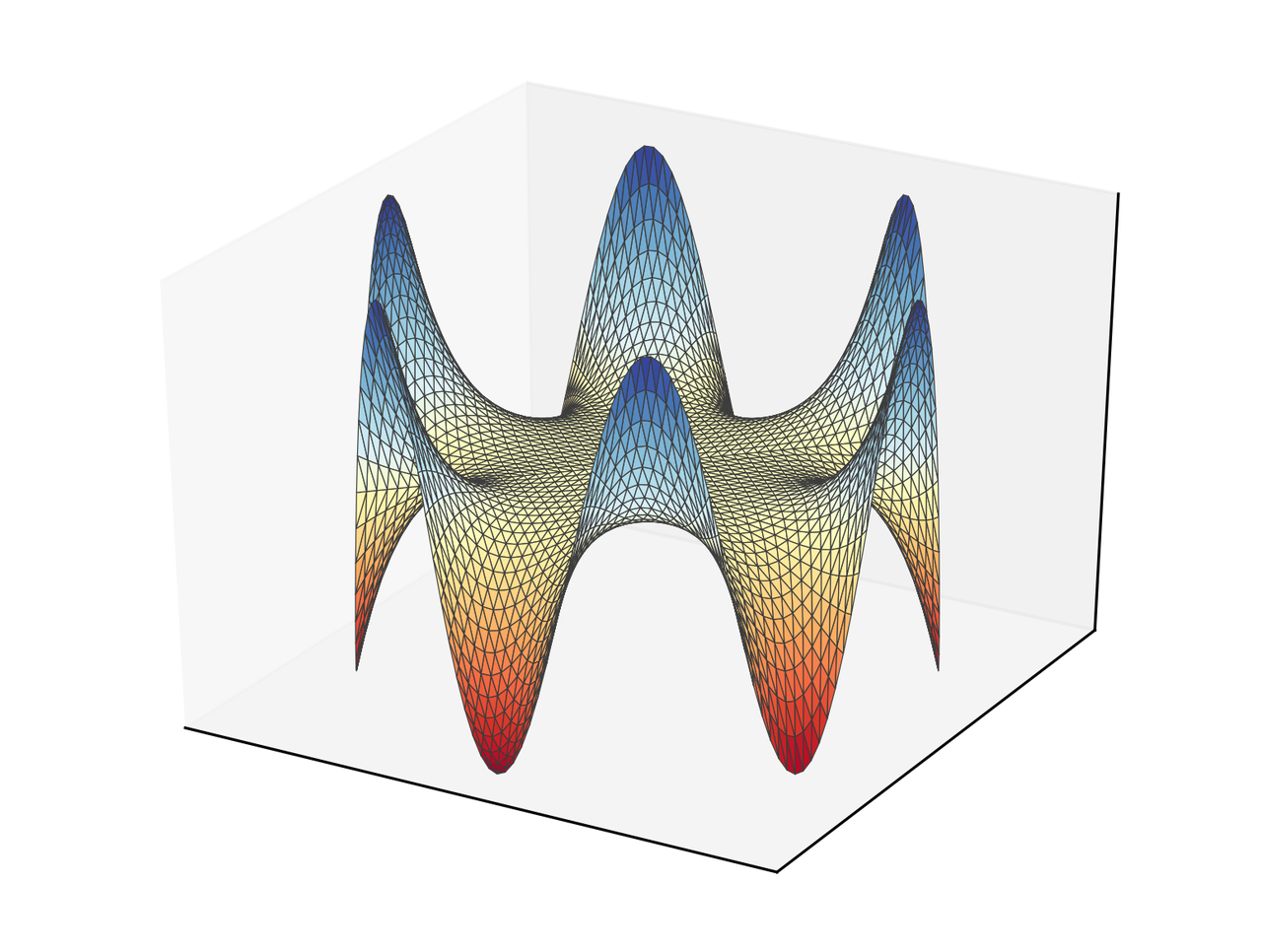
import numpy as np from numpy import log, arctan2, pi, mean, arctan from numpy.linalg import norm, solve import matplotlib.pyplot as plt from mpl_toolkits.mplot3d import Axes3D import meshio def assem(coords, elems): """Assembly matrices for the BEM problem Parameters ---------- coords : ndarray, float Coordinates for the nodes. elems : ndarray, int Connectivity for the elements. Returns ------- Gmat : ndarray, float Influence matrix for the flow. Fmat : ndarray, float Influence matrix for primary variable. """ nelems = elems.shape[0] Gmat = np.zeros((nelems, nelems)) Fmat = np.zeros((nelems, nelems)) for ev_cont, elem1 in enumerate(elems): for col_cont, elem2 in enumerate(elems): pt_col = mean(coords[elem2], axis=0) if ev_cont == col_cont: L = norm(coords[elem1[1]] - coords[elem1[0]]) Gmat[ev_cont, ev_cont] = - L/(2*pi)*(log(L/2) - 1) Fmat[ev_cont, ev_cont] = - 0.5 else: Gij, Fij = influence_coeff(elem1, coords, pt_col) Gmat[ev_cont, col_cont] = Gij Fmat[ev_cont, col_cont] = Fij return Gmat, Fmat def influence_coeff(elem, coords, pt_col): """Compute influence coefficients Parameters ---------- elems : ndarray, int Connectivity for the elements. coords : ndarray, float Coordinates for the nodes. pt_col : ndarray Coordinates of the colocation point. Returns ------- G_coeff : float Influence coefficient for flows. F_coeff : float Influence coefficient for primary variable. """ dcos = coords[elem[1]] - coords[elem[0]] dcos = dcos / norm(dcos) rotmat = np.array([[dcos[1], -dcos[0]], [dcos[0], dcos[1]]]) r_A = rotmat.dot(coords[elem[0]] - pt_col) r_B = rotmat.dot(coords[elem[1]] - pt_col) theta_A = arctan2(r_A[1], r_A[0]) theta_B = arctan2(r_B[1], r_B[0]) if norm(r_A) <= 1e-6: G_coeff = r_B[1]*(log(norm(r_B)) - 1) + theta_B*r_B[0] elif norm(r_B) <= 1e-6: G_coeff = -(r_A[1]*(log(norm(r_A)) - 1) + theta_A*r_A[0]) else: G_coeff = r_B[1]*(log(norm(r_B)) - 1) + theta_B*r_B[0] -\ (r_A[1]*(log(norm(r_A)) - 1) + theta_A*r_A[0]) F_coeff = theta_B - theta_A return -G_coeff/(2*pi), F_coeff/(2*pi) def eval_sol(ev_coords, coords, elems, u_boundary, q_boundary): """Evaluate the solution in a set of points Parameters ---------- ev_coords : ndarray, float Coordinates of the evaluation points. coords : ndarray, float Coordinates for the nodes. elems : ndarray, int Connectivity for the elements. u_boundary : ndarray, float Primary variable in the nodes. q_boundary : [type] Flows in the nodes. Returns ------- solution : ndarray, float Solution evaluated in the given points. """ npts = ev_coords.shape[0] solution = np.zeros(npts) for k in range(npts): for ev_cont, elem in enumerate(elems): pt_col = ev_coords[k] G, F = influence_coeff(elem, coords, pt_col) solution[k] += u_boundary[ev_cont]*F - q_boundary[ev_cont]*G return solution #%% Simulation mesh = meshio.read("disk.msh") elems = mesh.cells["line"] bound_nodes = list(set(elems.flatten())) coords = mesh.points[bound_nodes, :2] x, y = coords.T x_m, y_m = 0.5*(coords[elems[:, 0]] + coords[elems[:, 1]]).T theta = np.arctan2(y_m, x_m) u_boundary = 3*np.cos(6*theta) #%% Assembly Gmat, Fmat = assem(coords, elems) #%% Solution q_boundary = solve(Gmat, Fmat.dot(u_boundary)) #%% Evaluation ev_coords = mesh.points[:, :2] ev_x, ev_y = ev_coords.T solution = eval_sol(ev_coords, coords, elems, u_boundary, q_boundary) #%% Visualization tris = mesh.cells["triangle"] fig = plt.figure() ax = fig.add_subplot(111, projection='3d') ax.plot_trisurf(ev_x, ev_y, solution, cmap="RdYlBu", lw=0.3, edgecolor="#3c3c3c") plt.xticks([]) plt.yticks([]) ax.set_zticks([]) plt.savefig("bem_solution.png", bbox_inches="tight", transparent=True, dpi=300)
The result in this case is the following.
Comments
Comments powered by Disqus